Sequence to function mapping of condensate proteomes (B3)
Objective
The goal of of B3 is to understand how condensate proteomes and their functions evolve. We aim to decipher which sequence perturbations can lead to gain/loss of condensates and tune specific molecular inter-actions within condensates in evolution and disease.
Project Description
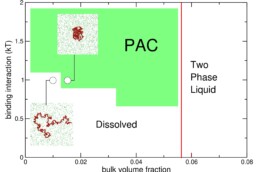
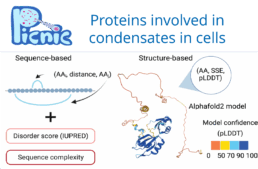
Extensive research on condensates focused on identifying their components, material proper-ties, and function. However, we still lack understanding of the mechanisms that target proteins into condensates and how condensates emerged during evolution. Building on the hypothesis that localization into condensates is encoded in protein sequences, we aim to advance the sequence-function mapping of condensate forming proteins and specifically intrinsically disordered regions (IDRs) that are common in condensates. Here, we will study not only genetic mutations but also phenotypic mutations that occur via transcription and translation errors, which we study in the Toth-Petroczy group. In collaboration with the Hyman group we built a condensate protein database that revealed wide-spread occurrence of condensates across the tree of life. Based on this curated data, we developed a machine learning algorithm to predict condensate proteins in any organism. Further, we have designed a suit of alignment-free algorithms to assess homology between unalignable IDR sequences (SHARK-dive) and developed a method to identify conserved motifs within a set of homologous IDR sequences (SHARK-capture). These tools allow us to address the origin and evolution of condensates and their proteomes, as well as to identify conserved and functional sequence features.
Research questions
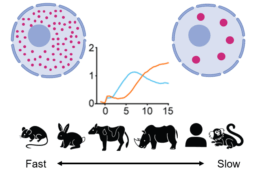
- How do condensate proteomes change in evolution?
- How do sequence perturbations impact condensate formation?
Thesis Project Topic
The evolution of condensate proteomes
Training
The PhD students will be trained in data science, coding, machine learning, statistics, and image analysis, mass spectrometry, molecular biology, biochemistry and biophysics techniques in collaborations with experimental groups.
Profile of Prospective Students
- Candidates have a Masters degree in biology, data science, or related fields
- Candidates should have a sound basis in computational or evolutionary biology, or closely related fields.
- Experience in coding and omics data analysis are expected

Supervisor: Agnes Toth-Petroczy
Protein plasticity and evolution
Discipline: Computational Biology
Affiliation: MPI-CBG | CSBD | Physics of Life (TU Dresden)
Contact: tothpet (at) mpi-cbg (dot) de
Explore other RTG Thesis Projects
Collaborations within the RTG
Click on the different project numbers (e.g. A1) to find out more about the theme of their ongoing collaborations and explore the project details
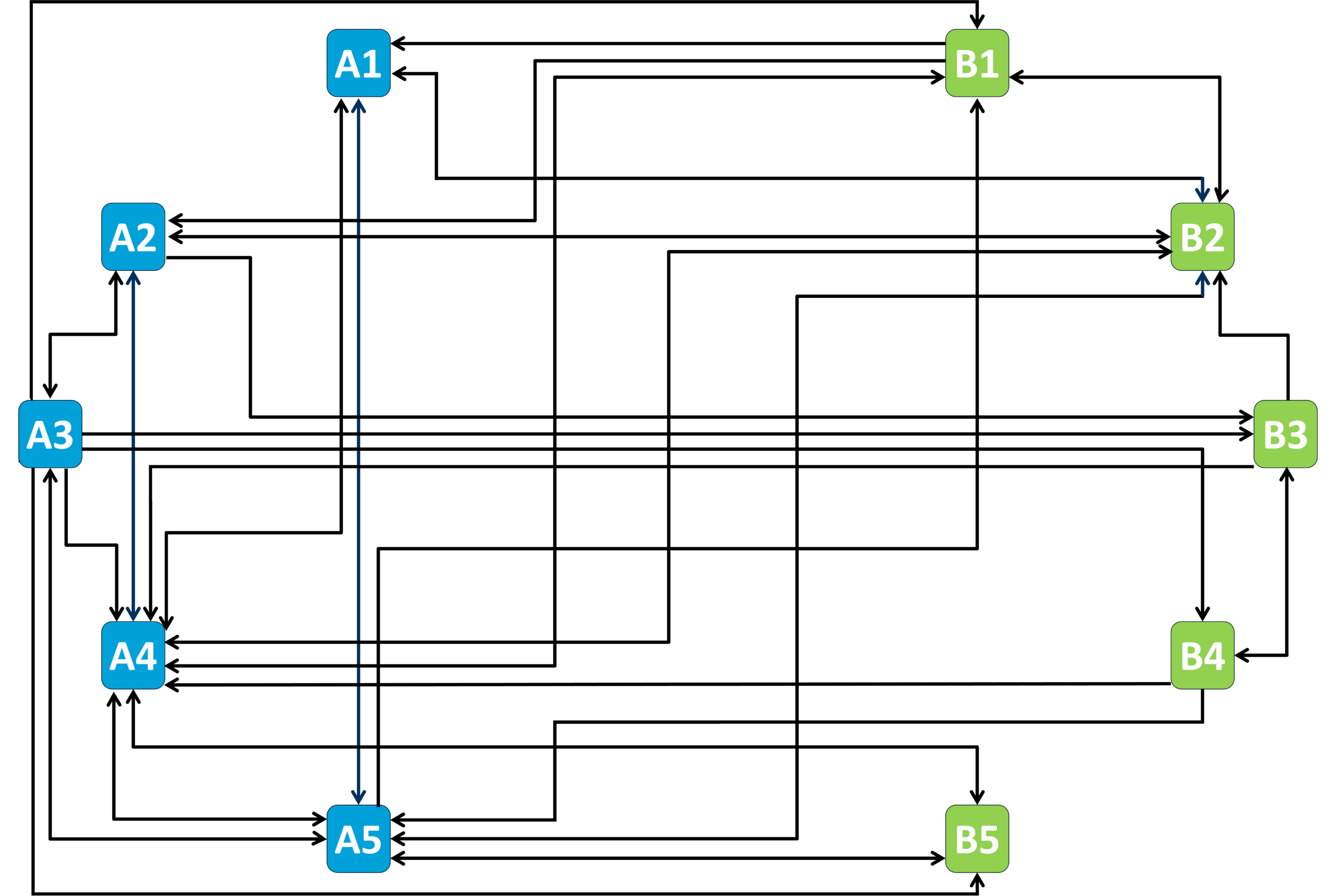
A2 - Biomolecular condensate regulation (Harmon)
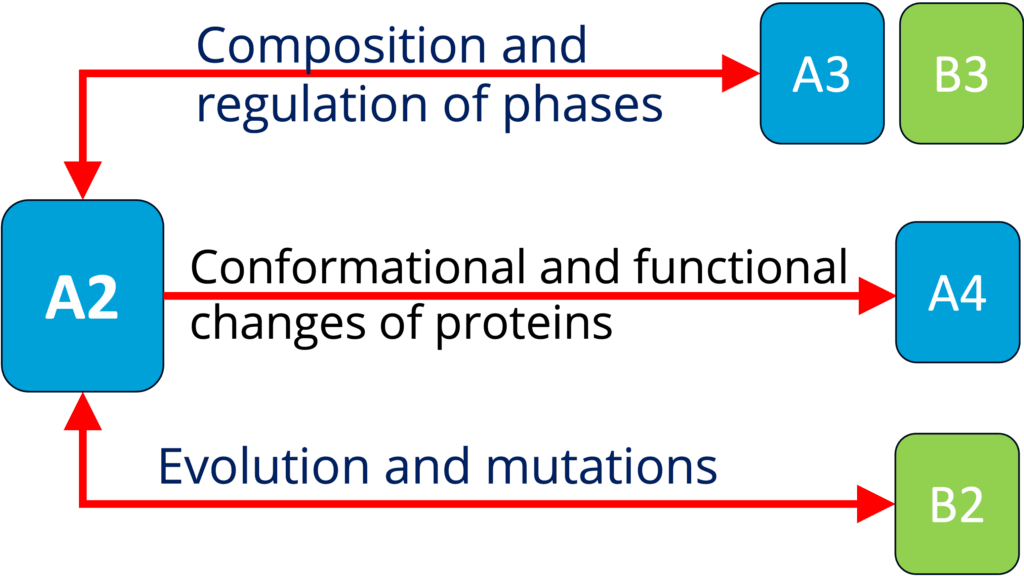
See project details: https://dresdencondensates.org/projects/a2/
A4 - Theory and simulation of polymer-assisted condensates (Sommer)
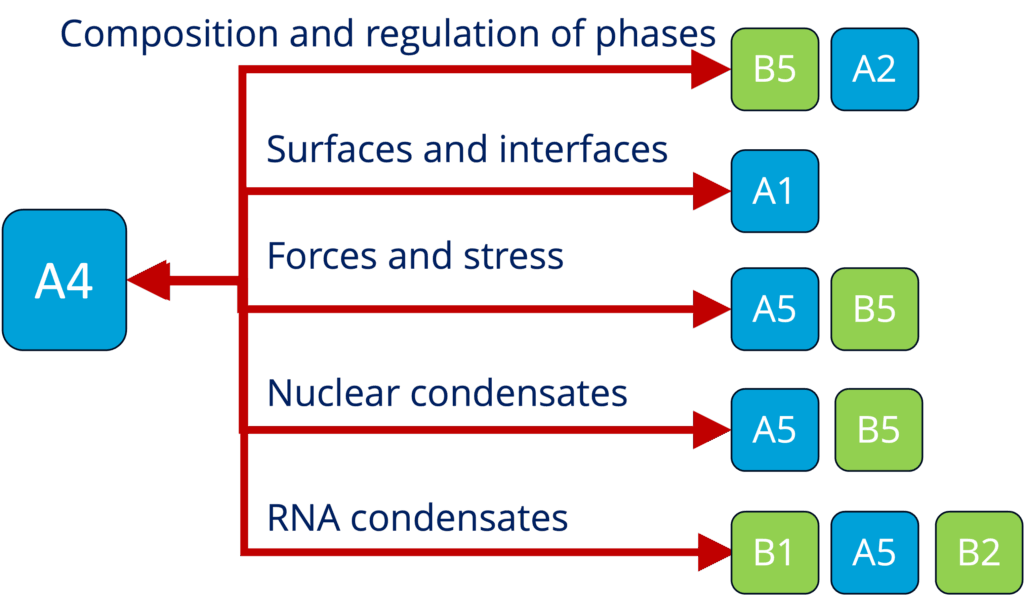
See project details: https://dresdencondensates.org/projects/a4/
B2 - Characterizing the role of RNP granules in ALS (Sterneckert)
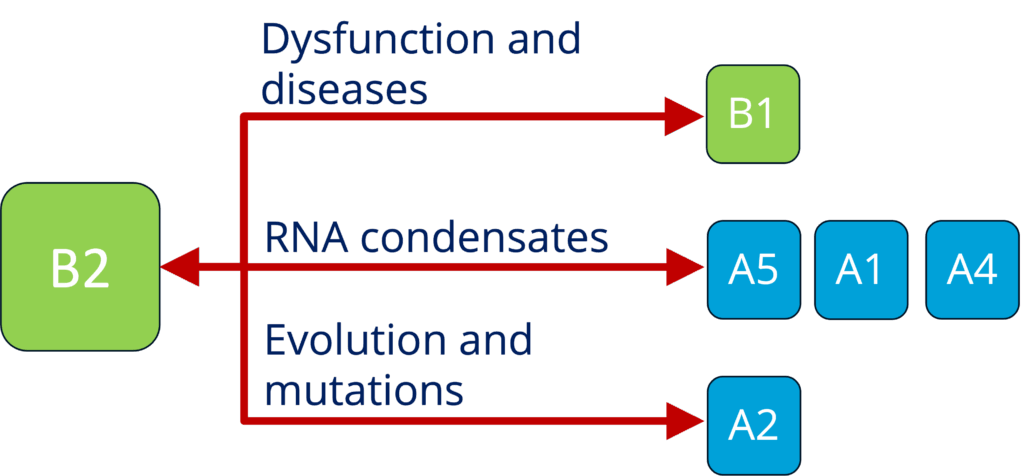
See project details: https://dresdencondensates.org/projects/b2/
A1 - Role of surface condensation for the assembly of cortical proteins (Honigmann)
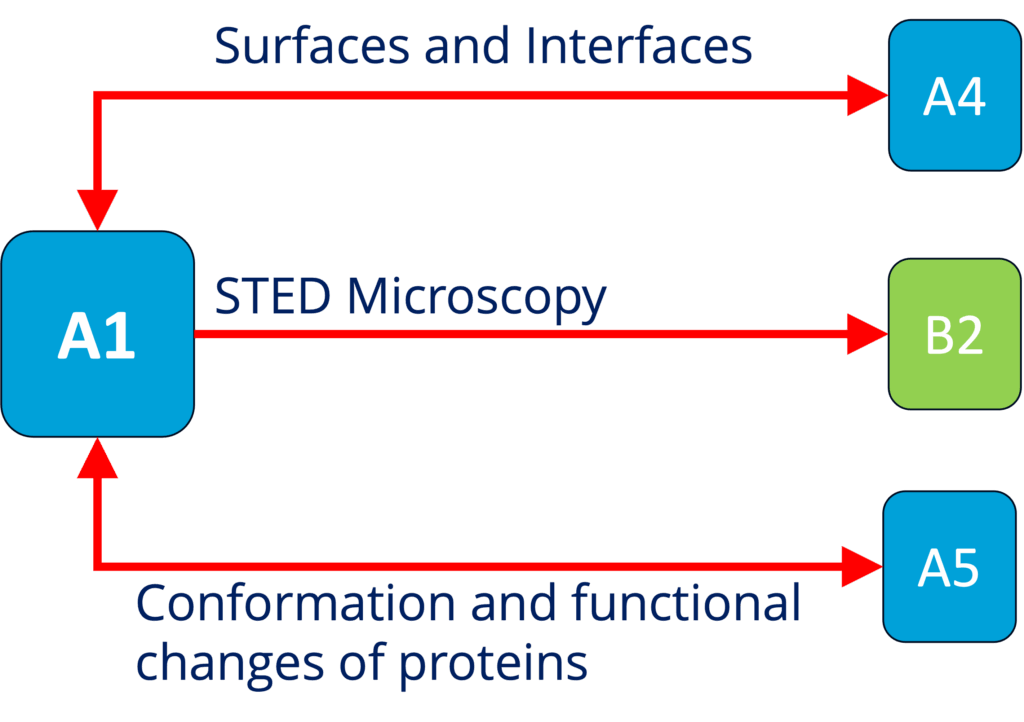
See project details: https://dresdencondensates.org/projects/a1/
A3 - Spectroscopy and local interactions in condensates and organization of the cytoplasm (Adams)
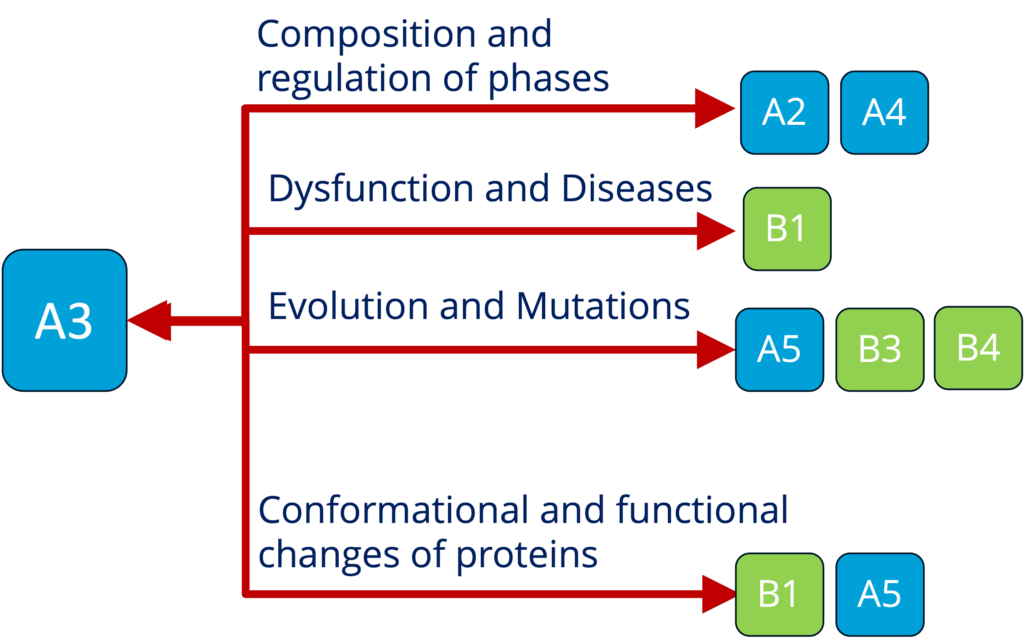
See project details: https://dresdencondensates.org/projects/a3/
A5 - Capillary forces and the force response of condensates (Jahnel and Grill)
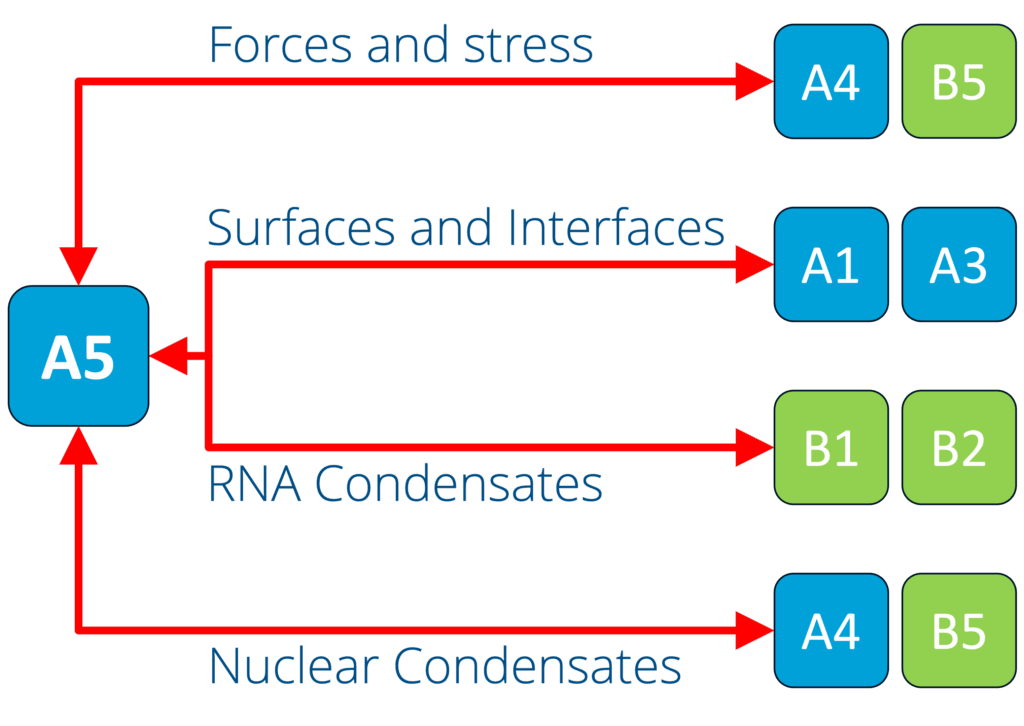
See project details: https://dresdencondensates.org/projects/a5/
B1 - Elucidating the mechanisms underlying mRNA translation regulation by condensation (biophysics and biochemistry) (Alberti and Schlierf)
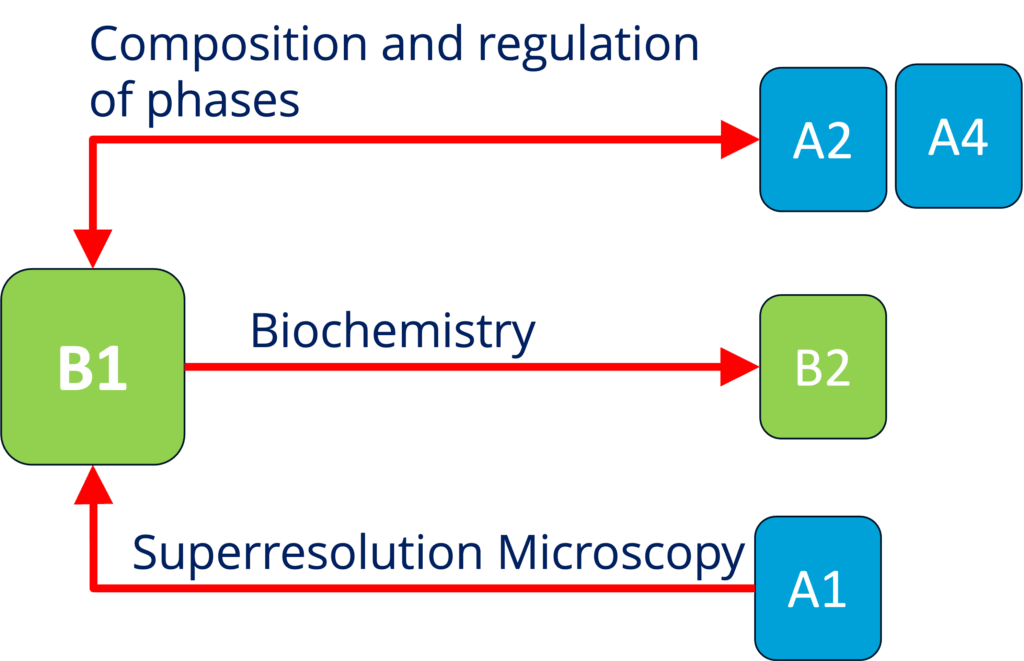
See project details: https://dresdencondensates.org/projects/b1/
B3 - Sequence to function mapping of condensate proteomes (Toth-Petroczy)
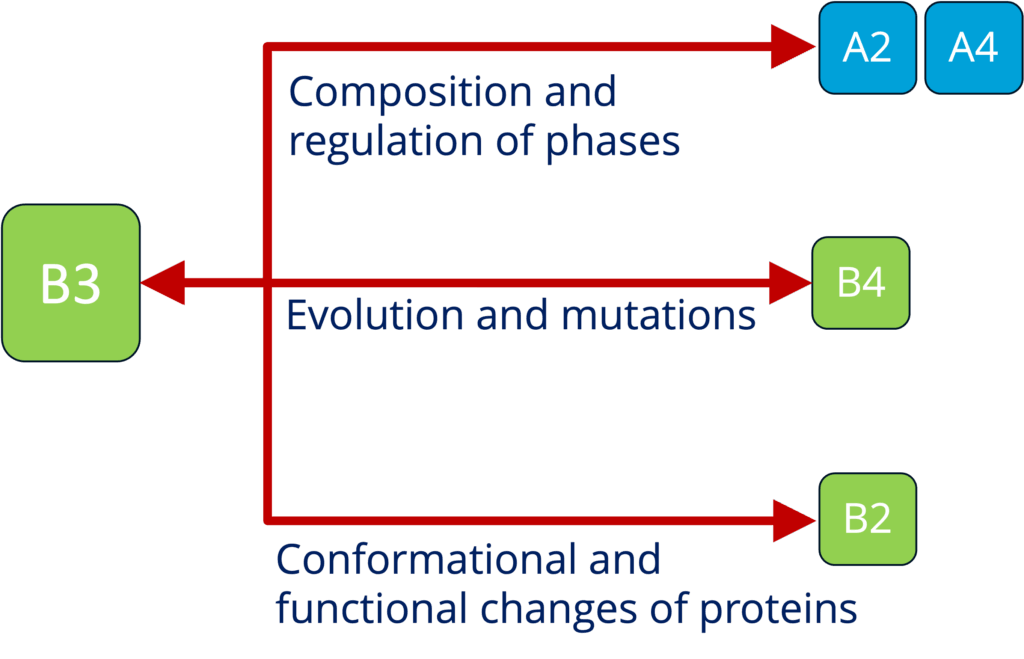
See project details: https://dresdencondensates.org/projects/b3/
B4 - Role of condensates in biological time across mammals (Ebisuya and Hyman)
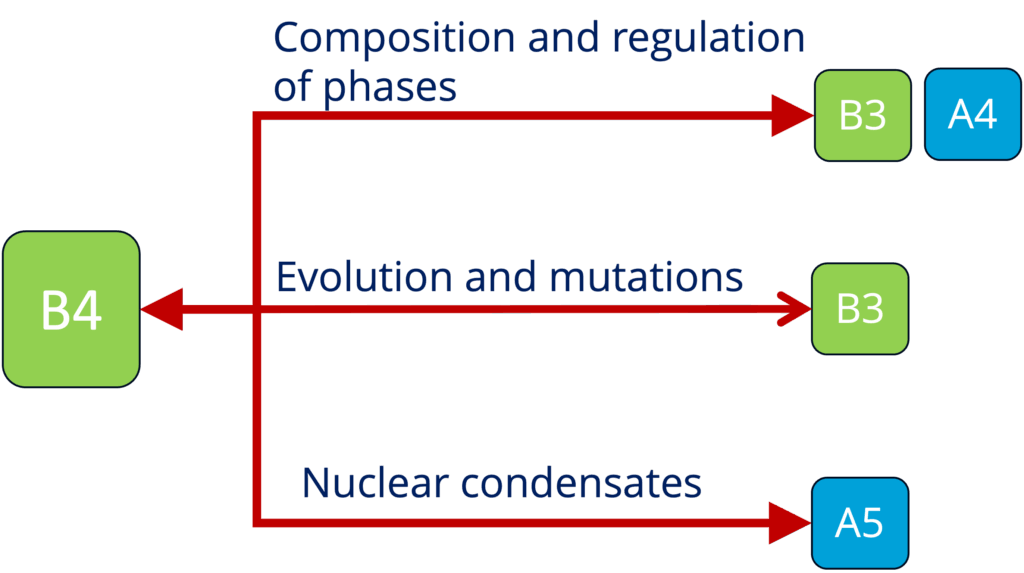
See project details: https://dresdencondensates.org/projects/b4/
B5 - Role of condensates in epigenetics (experiments and theory) (Brugués and Schiessel)
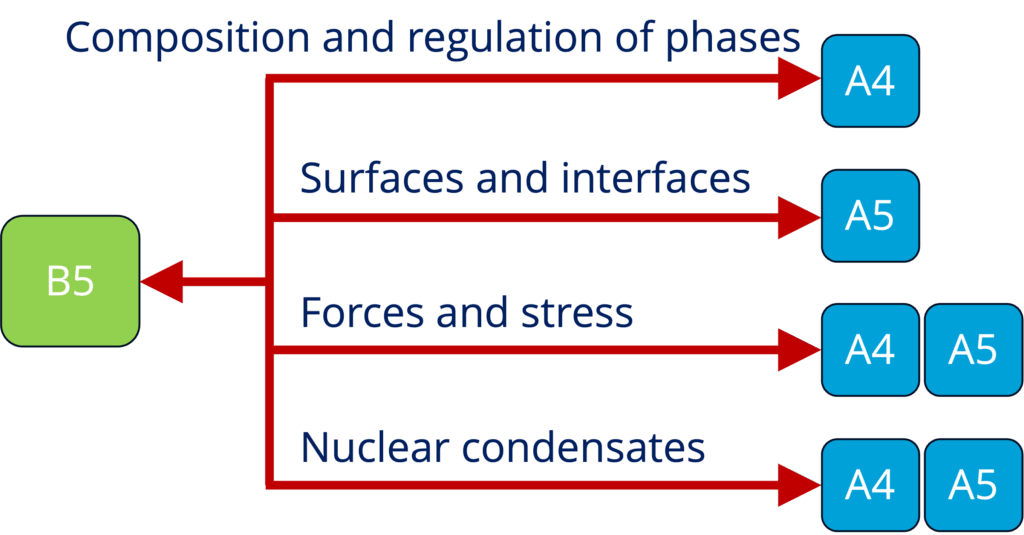
See Project Details: https://dresdencondensates.org/projects/b5/